DataFrames - Part 1
Last updated on 2024-09-07 | Edit this page
Estimated time: 120 minutes
Download Chapter notebook (ipynb)
Mandatory Lesson Feedback Survey
Overview
Questions
- What is a Dataframe, and how can we read data into one?
- What are the different methods for manipulating data in Dataframes?
- What makes data visualisation simple, in Python?
Objectives
- Import a dataset as a Pandas Dataframe
- Inspect a Dataframe and access data
- Produce an overview of data features
- Create data plots using Matplotlib
Prerequisites
- Indexing of arrays
- For-loop through an array
- Basic statistics (distributions, mean, median and standard deviation)
The diabetes data set is one of the challenging task.
Challenge: The diabetes dataset
Here is a screenshot of a diabetes dataset. It is taken from this webpage, and is one of the example datasets used to illustrate machine learning functionality in scikit-learn (Part II of the L2D course).
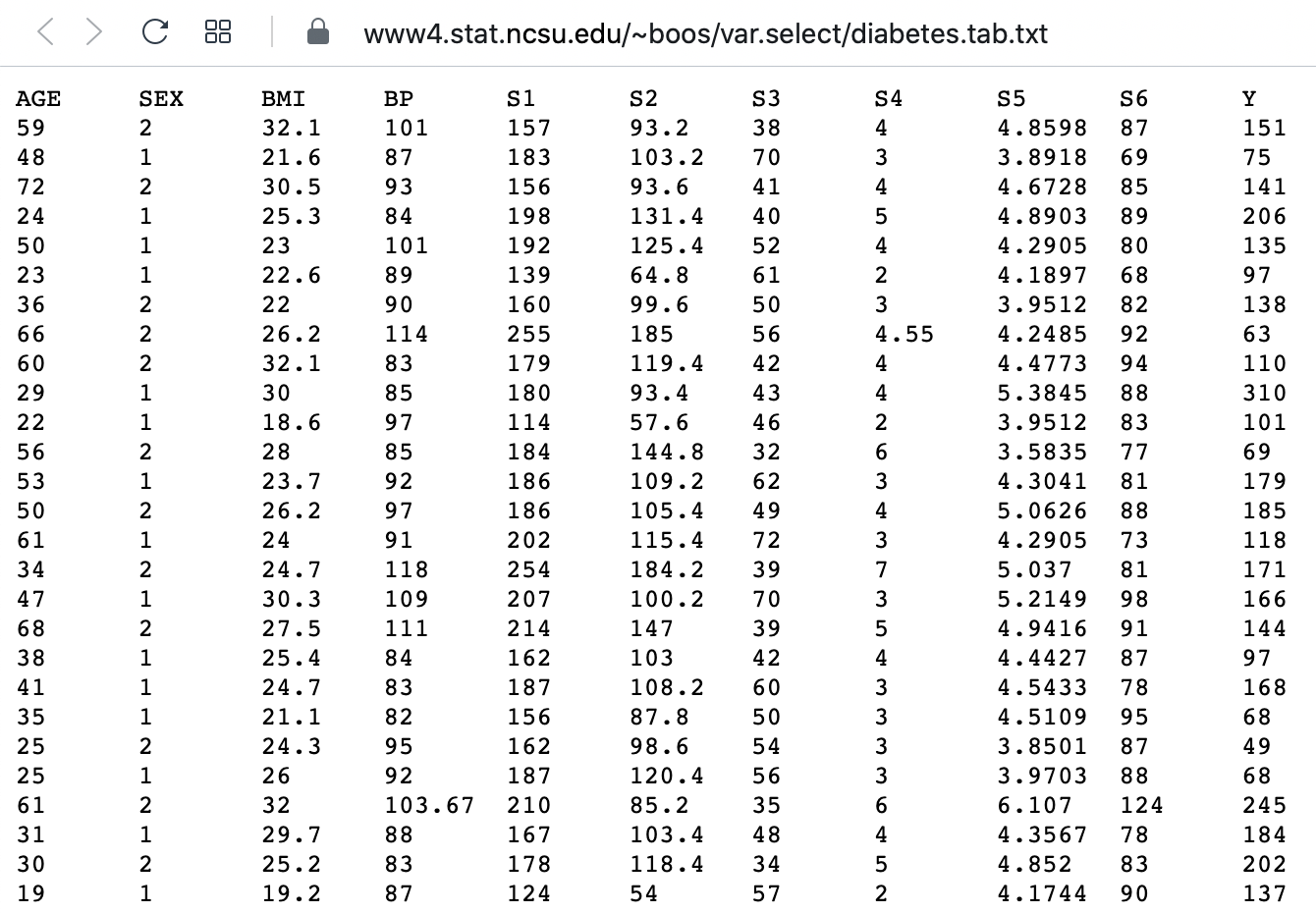
This figure captures only the top part of the data. On the webpage, you will need to scroll down considerably to view all of it. Thus, our first data science task, will be to obtain an overview of this datset.
The lesson
- Introduces code to read and inspect the data
- Works with a specific Dataframe and explains methods used to get an overview of the data
- Discusses the concept of ‘distribution’ as a way of summarising data within a single figure
To familiarise yourself with a dataset you need to:
- Access the data
- Check the content
- Produce a summary of basic properties
In this lesson we will look solely at univariate features, where each data columns are studied independently of the others in the datasets. Further properties and bivariate features will be the topic of the next lesson.
Work-Through Example
Reading data into a Pandas DataFrame
The small practice data file for this section is called
‘everleys_data.csv’, and can be downloaded using the link given above in
Summary and Setup for this Lesson. To start,
please create a subfolder called ‘data’ in the current directory and put
the data file in it. It can now be accessed using the relative path
data/everleys_data.csv or
data\everleys_data.csv, respectively.
The file everleys_data.csv contains serum concentrations of
calcium and sodium ions, sampled from 17 patients with Everley’s
syndrome - a rare genetic disorder that results in sufferers
experiencing developmental delays, intellectual and physical
abnormalities. The data are taken from a BMJ
statistics tutorial, and are stored as comma-separated values (csv):
with two values given for each patient.
To get to know a dataset, we will use the Pandas package and the Matplotlib plotting library. The Pandas package for data science is included in the Anaconda distribution of Python. Check this link for installation instructions to get started.
If you are not using the Anaconda distribution, please refer to these guidelines.
In order to use the functions contained in Pandas, they must first to be
imported. Since our dataset is in a ‘.csv’ file, we must first read it
from a csv file. For this, we must import the function
read_csv, which will create a Pandas DataFrame
from data provided in a ‘.csv’ file.
Executing this code does not lead to any output on the screen. However, the function is now ready to be used. To use it, we type its name and provide the required arguments. The following code should import the Everley’s data into your Python Notebook:
PYTHON
# For Mac OS X and Linux:
# (please go to the next cell if using Windows)
df = read_csv("data/everleys_data.csv")
Note the orientation of backward and forward slashes that differentiate
filepaths given between Unix-based systems, and Windows. This code uses
the read_csv function from Pandas to read data from a data
file, in this case a file with extension ‘.csv’. Note that the location
of the data file is specified within quotes by the relative path to the
subfolder ‘data’, followed by the file name. Use your file browser or
the browser in JupyterLab (or an ‘Explorer’-type pane in your IDE of
choice) to check that subfolder does indeed exists, and contains the
file within it.
After execution of the code, the data are contained in a variable
called df. This is a structure referred to as a Pandas
DataFrame.
A Pandas DataFrame is a 2-dimensional labelled data structure, with columns of (potentially different) types. You can think of it as a spreadsheet.
To see the contents of df, simply use:
OUTPUT
calcium sodium
0 3.455582 112.690980
1 3.669026 125.663330
2 2.789910 105.821810
3 2.939900 98.172772
4 5.426060 97.931489
5 0.715811 120.858330
6 5.652390 112.871500
7 3.571320 112.647360
8 4.300067 132.031720
9 1.369419 118.499010
10 2.550962 117.373730
11 2.894129 134.052390
12 3.664987 105.346410
13 1.362779 123.359490
14 3.718798 125.021060
15 1.865868 112.075420
16 3.272809 117.588040
17 3.917591 101.009870(Compare with the result of print(df) which displays the
contents in a different format.)
The output shows in the first column an index, integers from 0 to 17; and the calcium and sodium concentrations in columns 2 and 3, respectively. The default indexing starts from zero (Python is a ‘zero-based’ programming language).
In a DataFrame, the first column is referred to as Indices, the first row is referred to as Labels. Note that the row with the labels is excluded from the row count. Similarly, the row with the indices is excluded from the column count.
For large datasets, the function head is a convenient
way to get a feel of the dataset.
OUTPUT
calcium sodium
0 3.455582 112.690980
1 3.669026 125.663330
2 2.789910 105.821810
3 2.939900 98.172772
4 5.426060 97.931489
Without any input argument, this displays the first five data lines of
data i the newly-created DataFrame. You can specify and alter the number
of rows displayed by including a single integer as argument,
e.g. head(10).
If you feel there are too many decimal places in the default view, you
can restrict their number by using the round method. The
numerical argument that you provide in the round parentheses controls
the number of decimal places the method rounds to, with digits up to 5
being rounded down, and above (and inclusive of) 5, being rounded up:
OUTPUT
calcium sodium
0 3.46 112.69
1 3.67 125.66
2 2.79 105.82
3 2.94 98.17
4 5.43 97.93While it is possible to see how many rows there are in a DataFrame by displaying the whole DataFrame and looking at the last index, there is a convenient way to obtain this number, directly:
OUTPUT
DataFrame has 18 rowsYou could see above, that the columns of the DataFrame have labels. To see all labels:
OUTPUT
Index(['calcium', 'sodium'], dtype='object')Now we can count the labels to obtain the number of columns:
OUTPUT
DataFrame has 2 columns
And if you want to have both the number of the rows and the number
columns displayed together, you can use the shape method.
Shape returns a tuple of two numbers: the first is the number of rows,
and the second is the number of columns.
OUTPUT
DataFrame has 18 rows and 2 columnsNotice that shape (like columns) is not
followed by round parentheses. It is not a function that can take
arguments. Technically, shape is a ‘property’ of the
DataFrame.
To find out what data type is contained in each of the columns, us
dtypes, another ‘property’:
OUTPUT
calcium float64
sodium float64
dtype: objectIn this case, both columns contain floating point (decimal) numbers.
Practice Exercise 1
Read data into a DataFrame
Download the data file ‘loan_data.csv’ using the link given above in Summary and Setup for this Lesson”. It contains data that can be used for the assessment of loan applications. Read the data into a DataFrame. It is best to assign it a name other than ‘df’ (to avoid overwriting the Evereley dataset).
Display the first ten rows of the Loan dataset to see its contents. It is taken from a tutorial on Data Handling in Python which you might find useful for further practice.
From this exercise we can see that a DataFrame can contain different types of data: real numbers (e.g. LoanAmount), integers (ApplicantIncome), categorical data (Gender), and strings (Loan_ID).
PYTHON
from pandas import read_csv
# dataframe from .csv file
df_loan = read_csv("data/loan_data.csv")
# display contents
df_loan.head(10)OUTPUT
Loan_ID Gender Married ... Loan_Amount_Term Credit_History Property_Area
0 LP001015 Male Yes ... 360.0 1.0 Urban
1 LP001022 Male Yes ... 360.0 1.0 Urban
2 LP001031 Male Yes ... 360.0 1.0 Urban
3 LP001035 Male Yes ... 360.0 NaN Urban
4 LP001051 Male No ... 360.0 1.0 Urban
5 LP001054 Male Yes ... 360.0 1.0 Urban
6 LP001055 Female No ... 360.0 1.0 Semiurban
7 LP001056 Male Yes ... 360.0 0.0 Rural
8 LP001059 Male Yes ... 240.0 1.0 Urban
9 LP001067 Male No ... 360.0 1.0 Semiurban
[10 rows x 12 columns]Accessing data in a DataFrame
If a datafile is large and you only want to check the format of data in a specific column, you can limit the display to that column. To access data contained in a specific column of a DataFrame, we can use a similar convention as in a Python dictionary, treating the column names as ‘keys’. E.g. to show all rows in column ‘Calcium’, use:
OUTPUT
0 3.455582
1 3.669026
2 2.789910
3 2.939900
4 5.426060
5 0.715811
6 5.652390
7 3.571320
8 4.300067
9 1.369419
10 2.550962
11 2.894129
12 3.664987
13 1.362779
14 3.718798
15 1.865868
16 3.272809
17 3.917591
Name: calcium, dtype: float64To access individual rows of a column we use two pairs of square brackets:
OUTPUT
0 3.455582
1 3.669026
2 2.789910
Name: calcium, dtype: float64Here all rules for slicing can be applied. As for lists and tuples, the indexing of rows is semi-inclusive, with the lower boundary included and upper boundary excluded. Note that the first pair of square brackets refers to columns, and the second pair refers to the rows. However, this is different from accessing items in a nested list, for instance.
Accessing items in a Pandas DataFrame is analogous to accessing the values in a Python dictionary by referring to its keys.
To access non-contiguous elements, we use an additional pair of square brackets (as if for a list within a list):
OUTPUT
1 3.669026
3 2.939900
7 3.571320
Name: calcium, dtype: float64
Another method for indexing and slicing a DataFrame is to use the
‘index location’ or iloc property. Note that properties in
Python differ from methods. Syntactically, they use the same dot
notation we are accustomed to with methods, but they differ in their use
of square brackets, rather than the round parentheses that methods
operate with. A property also refers directly to a specific
attribute of an object.
iloc refers first to the rows of data, and
then to columns - by index; all contained within a single pair of
brackets. For example, to obtain all the rows of the first column (index
0), you use:
OUTPUT
0 3.455582
1 3.669026
2 2.789910
3 2.939900
4 5.426060
5 0.715811
6 5.652390
7 3.571320
8 4.300067
9 1.369419
10 2.550962
11 2.894129
12 3.664987
13 1.362779
14 3.718798
15 1.865868
16 3.272809
17 3.917591
Name: calcium, dtype: float64To display only the first three calcium concentrations, slicing is used: note that the upper boundary is excluded):
OUTPUT
0 3.455582
1 3.669026
2 2.789910
Name: calcium, dtype: float64To access non-consecutive values, we can use a pair of square brackets within the outer pair of square brackets:
OUTPUT
2 2.78991
4 5.42606
7 3.57132
Name: calcium, dtype: float64Similarly, we can access values from multiple columns:
OUTPUT
calcium sodium
2 2.78991 105.821810
4 5.42606 97.931489
7 3.57132 112.647360To pick only the even rows from the two columns, note the following colon notation:
OUTPUT
calcium sodium
0 3.455582 112.690980
2 2.789910 105.821810
4 5.426060 97.931489
6 5.652390 112.871500
8 4.300067 132.031720
10 2.550962 117.373730
12 3.664987 105.346410
14 3.718798 125.021060
16 3.272809 117.588040The number after the second colon indicates the stepsize.
Practice Exercise 2
Select data from DataFrame
Display the calcium and sodium concentrations of all patients - except the first. Check the model solution at the bottom for options.
OUTPUT
calcium sodium
1 3.669026 125.663330
2 2.789910 105.821810
3 2.939900 98.172772
4 5.426060 97.931489
5 0.715811 120.858330
6 5.652390 112.871500
7 3.571320 112.647360
8 4.300067 132.031720
9 1.369419 118.499010
10 2.550962 117.373730
11 2.894129 134.052390
12 3.664987 105.346410
13 1.362779 123.359490
14 3.718798 125.021060
15 1.865868 112.075420
16 3.272809 117.588040
17 3.917591 101.009870Mixing the different methods of accessing specific data in a DataFrame can be confusing, and requires practice and diligence.
Search for missing values
Some tables contain missing entries. You can check a DataFrame for
such missing entries. If no missing entry is found, the function
isnull will return False.
OUTPUT
calcium False
sodium False
dtype: boolThis shows that there are no missing entries in our DataFrame.
Practice Exercise 3
Find NaN in DataFrame
In the Loan dataset, check the entry ‘Self-employed’ for ID LP001059. It shows how a missing value is represented as ‘NaN’ (not a number).
Verify that the output of isnull in this case is
True
Basic data features:
Summary Statistics
To get a summary of basic data features, it is possible to use the
function describe:
OUTPUT
calcium sodium
count 18.000000 18.000000
mean 3.174301 115.167484
std 1.306652 10.756852
min 0.715811 97.931489
25% 2.610699 107.385212
50% 3.364195 115.122615
75% 3.706355 122.734200
max 5.652390 134.052390
The describe function produces a new DataFrame (here called
‘description’) that contains the number of samples, the mean, the
standard deviation, 25th, 50th, 75th percentile, and the minimum and
maximum values for each column of the data. Note that the indices of the
rows have now been replaced by strings. To access rows, it is possible
to refer to those names using the loc property. Thus, in
order to access the mean of the calcium concentrations from the
description, each of the following is valid:
PYTHON
# Option 1
description.loc['mean']['calcium']
# Option 2
description.loc['mean'][0]
# Option 3
description['calcium']['mean']
# Option 4
description['calcium'][1]OUTPUT
3.1743005405555555
3.1743005405555555
3.1743005405555555
3.1743005405555555Practice Exercise 4
Use your own .csv dataset to practice. (If you don’t have a dataset at
hand, any excel table can be exported as .csv.) Firstly, read it into a
DataFrame, and proceed by checking its header, accessing individual
values or sets of values etc. Create a statistical summary using
describe, and check for missing values using
.isnull.
[ad libitum]
Iterating through the columns
Now we know how to access all data in a DataFrame and how to get a statistical summary statistics over each column.
Here is code to iterate through the columns and access the first two concentrations:
OUTPUT
0 3.455582
1 3.669026
Name: calcium, dtype: float64
0 112.69098
1 125.66333
Name: sodium, dtype: float64As a slightly more complex example, we access the median (‘50%’) of each column in the description, and add it to a list:
PYTHON
conc_medians = list()
for col in df:
conc_medians.append(df[col].describe()['50%'])
print('The columns are: ', list(df.columns))
print('The medians are: ', conc_medians)OUTPUT
The columns are: ['calcium', 'sodium']
The medians are: [3.3641954, 115.122615]This approach is useful for DataFrames with a larger number of columns. For instance, it is possible to follow this by creating a boxplot or histogram for the means, medians etc. of the DataFrame, thus giving a comprehensive overview of all (comparable) columns.
Selecting a subset based on a template
Often, an analysis of a dataset may required on only part of the data. This can often be formulated by using a logical condition which specifies the required subset.
For this we will assume that some of the data are labelled ‘0’ and some are labelled ‘1’. Let us therefore see how to add a new column to our Evereleys DataFrame, which contains the labels (which are, in this example, arbitrary).
Firstly, we can randomly create as many labels as we have rows in the DataFrame. We can use the randint function, which can be imported from the numpy.random module of the NumPy library. In its simplest form, the randint function accepts two arguments. Firstly, the upper bound of the integer needed, which defaults to zero. As Python is exclusive of the upper bound, providing ‘2’ will thus yield either ‘0’ or ‘1’ only.
PYTHON
from numpy.random import randint
no_rows = len(df)
randomLabel = randint(2, size=no_rows)
print('Number of rows: ', no_rows)
print('Number of Labels:', len(randomLabel))
print('Labels: ', randomLabel)OUTPUT
Number of rows: 18
Number of Labels: 18
Labels: [1 0 1 0 1 0 0 1 1 0 0 1 1 0 0 1 1 0]Note how we obtain the number of rows (18) using len function and do not explicitly state it in the code.
Next, we must create a new data column in our df DataFrame
which contains the labels. In order to create a new column, you may
simply refer to a column name that does not yet exist, and subsequently
assign values to it. Let us call it ‘gender’, assuming that ‘0’
represents male and ‘1’ represents female.
As gender specification can include more than two labels, try to create a column with more than two randomly assigned labels e.g. (0, 1, 2).
OUTPUT
calcium sodium gender
0 3.455582 112.690980 1
1 3.669026 125.663330 0
2 2.789910 105.821810 1
3 2.939900 98.172772 0
4 5.426060 97.931489 1Now we can use the information contained in ‘gender’ to filter the data by gender. To achieve this, we use a conditional statement. Let us check which of the rows are labelled as ‘1’:
OUTPUT
0 True
1 False
2 True
3 False
4 True
5 False
6 False
7 True
8 True
9 False
10 False
11 True
12 True
13 False
14 False
15 True
16 True
17 False
Name: gender, dtype: boolIf we assign the result of the conditional statement (a boolean: True or False) to a variable, then this variable can act as a template to filter the data. If we call the DataFrame with that variable, we will only get the rows where the condition was found to be True:
OUTPUT
calcium sodium gender
0 3.455582 112.690980 1
2 2.789910 105.821810 1
4 5.426060 97.931489 1
7 3.571320 112.647360 1
8 4.300067 132.031720 1
11 2.894129 134.052390 1
12 3.664987 105.346410 1
15 1.865868 112.075420 1
16 3.272809 117.588040 1Using the boolean, we only pick the rows that are labelled ‘1’ and thus get a subset of the data according to the label.
Practice Exercise 5
Using a template
Modify the code to calculate the number of samples labelled 0 and check the number of rows of that subset.
Visualisation of data
It is easy to see from the numbers that the concentrations of sodium are much higher than those of calcium. However, to incorporate comparisons of medians, percentiles and the spread of the data, it is better to use visualisation.
The simplest way to visualise data, is to use Pandas’ functionality which offers direct methods of plotting your data. Here is an example where a boxplot is created for each column:
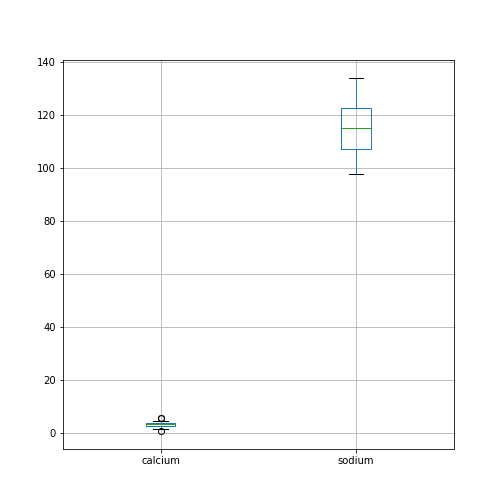
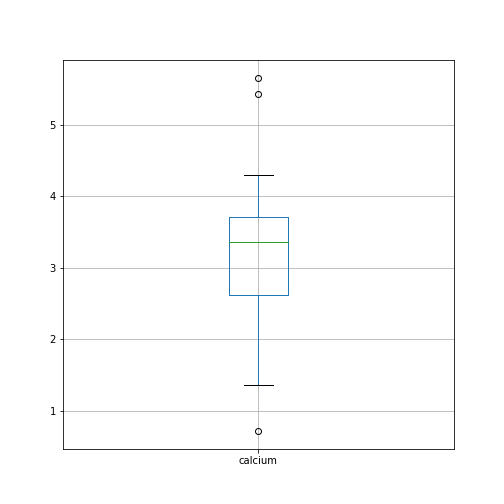
By default, boxplots are shown for all columns if no further argument is given to the function (empty round parentheses). As the calcium plot is quite condensed, we may wish to visualise it, separately. This can be done by specifying the calcium column as an argument:
Plots using Matplotlib
Matplotlib is a comprehensive library for creating static, animated and interactive visualizations in Python.
The above is an easy way to create boxplots directly on the DataFrame. It is based on the library Matplotlib and specifically uses the pyplot library. For simplicity, this code is put into a convenient Pandas function.
However, we are going to use Matplotlib extensively later on in the course, and we will therefore start by introducing a more direct and generic way of using it.
To do this, we import the function subplots from the pyplot library:
The way to use subplots is to first set up a figure environment (below referred to in the code as an object titled ‘fig’) and an empty coordinate system (below referred to as object ‘ax’). The plot is then created using one of the many methods available in Matplotlib. We will proceed by applying it to the coordinate system, ‘ax’.
As an example, let us create a boxplot of the calcium variable. As an argument of the function we need to specify the data. We can use the values of the ‘calcium’ concentrations from the column with the same name:
PYTHON
fig, ax = subplots()
ax.boxplot(df['calcium'])
ax.set_title('Boxplot of Everley Calcium')
show()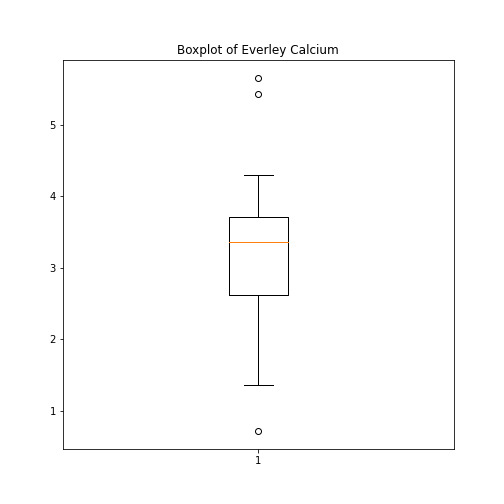
Note how we define the title of the plot by referring to the same
coordinate system ax.
The value of subplots becomes apparent when it is used to generate more than one plot as part of a single figure: one of its many useful features.
Here is an example whereby we create two boxplots adjacent to each other. The keyword arguments to use is ‘ncols’ which is the number of figures per row. ‘ncols=2’ indicates that you are plotting two plots adjacent to each other.
PYTHON
fig, ax = subplots(ncols=2)
ax[0].boxplot(df['calcium'])
ax[0].set_title('Calcium')
ax[1].boxplot(df['sodium'])
ax[1].set_title('Sodium');
show()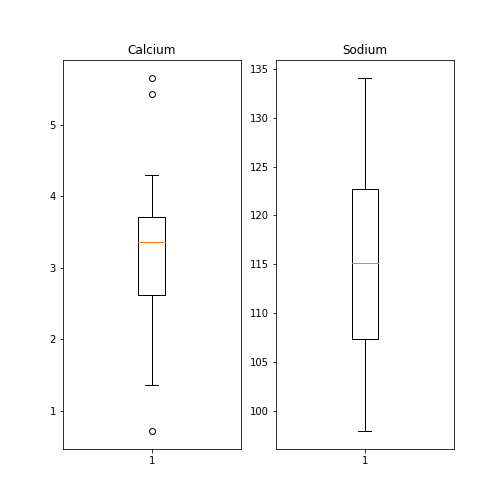
Each of these subplots must now be referred to using indexing the coordinate system ‘ax’. This figure gives a good overview of the Everley’s data.
If you prefer to have the boxplots of both columns in a single figure, this can also be done:
PYTHON
fig, ax = subplots(ncols=1, nrows=1)
ax.boxplot([df['calcium'], df['sodium']], positions=[1, 2])
ax.set_title('Boxplot of Calcium (left) and Sodium (right)')
show()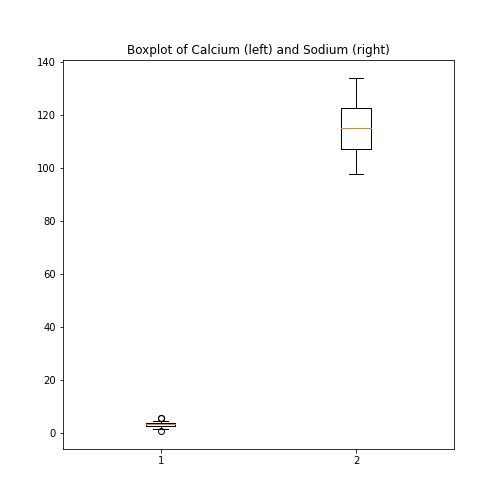
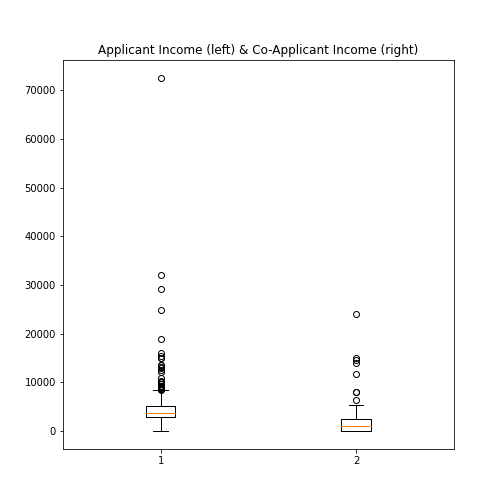
Practice Exercise 6
Boxplot from Loan data
Plot the boxplots of the ‘ApplicantIncome’ and the ‘CoapplicantIncome’ in the Loan data using the above code.
Histogram
Another good visual overview for data is the histogram. Containers or ‘bins’ are created over the range of values found within a column, and the count of the values for each bin is plotted on the vertical (y-)axis.
PYTHON
fig, ax = subplots(ncols=2, nrows=1)
ax[0].hist(df['calcium'])
ax[0].set(xlabel='Calcium', ylabel='Count');
ax[1].hist(df['sodium'])
ax[1].set(xlabel='Sodium', ylabel='Count');
fig.suptitle('Histograms of Everley concentrations', fontsize=15);
show()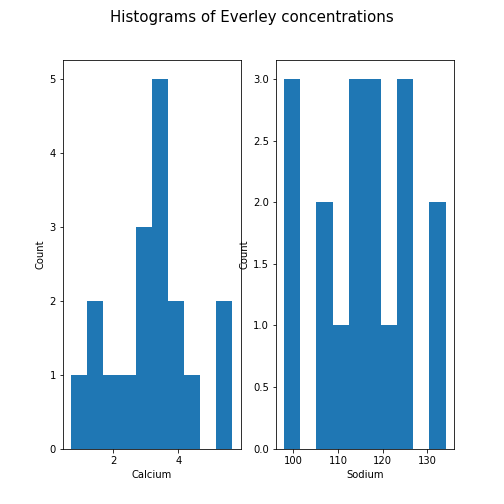
This example code also demonstrates how to use methods from within subplots to add labels to the axes, together with a title for the overall figure.
Unless specified, a default value is used for the generation of the bins. It is set to 10 bins over the range of which values are found. The number of bins in the histogram can be changed using the keyword argument ‘bins’:
PYTHON
fig, ax = subplots(ncols=2, nrows=1)
ax[0].hist(df['calcium'], bins=5)
ax[0].set(xlabel='Calcium, 5 bins', ylabel='Count');
ax[1].hist(df['calcium'], bins=15)
ax[1].set(xlabel='Calcium, 15 bins', ylabel='Count');
fig.suptitle('Histograms with Different Binnings', fontsize=16);
show()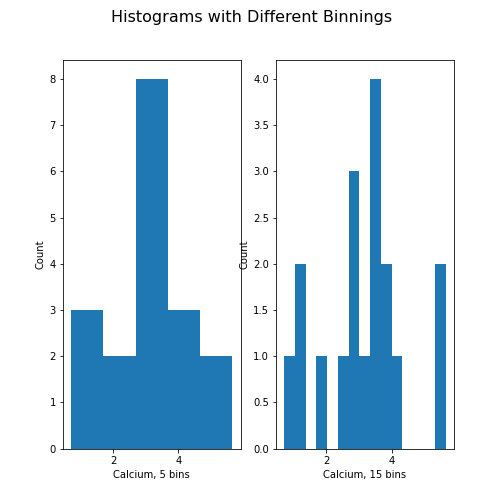
Note how the y-axis label of the right figure is slightly misplaced,
and overlapping the border of the left figure. In order to correct for
the placement of the labels and the title, you can use
tight_layout automatically adjust for this:
PYTHON
fig, ax = subplots(ncols=2, nrows=1)
ax[0].hist(df['calcium'], bins=5)
ax[0].set(xlabel='Calcium, 5 bins', ylabel='Count');
ax[1].hist(df['calcium'], bins=15)
ax[1].set(xlabel='Calcium, 15 bins', ylabel='Count');
fig.suptitle('Histograms with Different Binnings', fontsize=16);
fig.tight_layout()
show()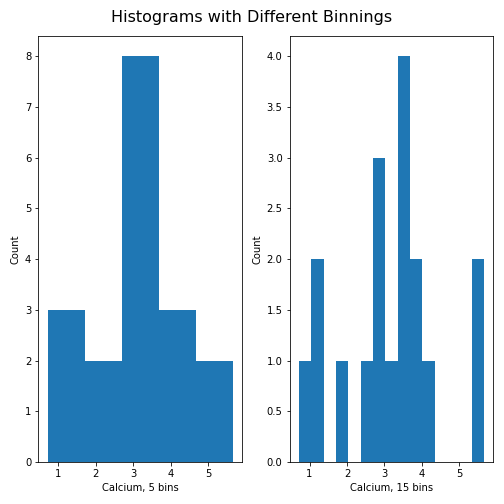
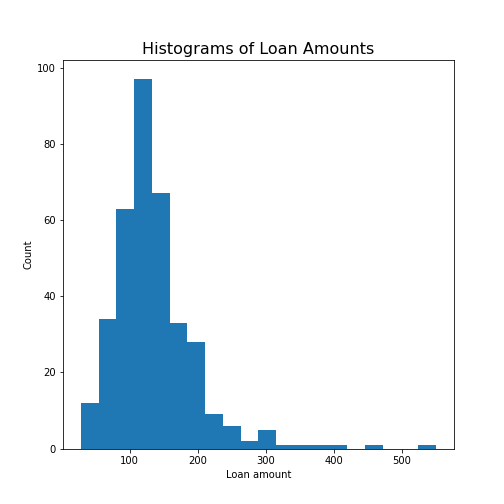
Practice Exercise 7:
Create the histogram of a column
Take the loan data and display the histogram of the loan amount that people asked for. (Loan amounts are divided by 1000, i.e. in k£ on the horizontal axis). Use 20 bins, as an example.
Handling the Diabetes Dataset
Let us return to the dataset that commenced our exploration of the handling of data within a DataFrame.
Next, we will:
- Import the diabetes data from ‘sklearn’
- Check the shape of the DataFrame and search for NANs
- Get a summary plot of one of its statistical quantities (i.e. mean) for all columns
Firstly, let’s import the dataset and check its head. In
some cases, this may take a moment: please be patient, and wait for the
numbers to appear as output below your code cell (if you’re using an
IDE).
PYTHON
from sklearn import datasets
diabetes = datasets.load_diabetes()
X = diabetes.data
from pandas import DataFrame
df_diabetes = DataFrame(data=X)
df_diabetes.head()OUTPUT
0 1 2 ... 7 8 9
0 0.038076 0.050680 0.061696 ... -0.002592 0.019907 -0.017646
1 -0.001882 -0.044642 -0.051474 ... -0.039493 -0.068332 -0.092204
2 0.085299 0.050680 0.044451 ... -0.002592 0.002861 -0.025930
3 -0.089063 -0.044642 -0.011595 ... 0.034309 0.022688 -0.009362
4 0.005383 -0.044642 -0.036385 ... -0.002592 -0.031988 -0.046641
[5 rows x 10 columns]If you don’t see all the columns, use the cursor to scroll to the right. Next, let’s check the number of columns and rows.
PYTHON
no_rows = len(df_diabetes)
no_cols = len(df_diabetes.columns)
print('Rows:', no_rows, 'Columns:', no_cols)OUTPUT
Rows: 442 Columns: 10There are 442 rows organised in 10 columns.
In order o obtain an overview, let us extract the mean of each column using the describe and plot all means as a bar chart. The Matplotlib function to plot a bar chart is called bar:
PYTHON
conc_means = list()
for col in df_diabetes:
conc_means.append(df_diabetes[col].describe()['mean'])
print('The columns are: ', list(df_diabetes.columns))
print('The medians are: ', conc_means, 2)OUTPUT
The columns are: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9]
The medians are: [-2.511816797794472e-19, 1.2307902309192911e-17, -2.2455642172282577e-16, -4.7975700837874414e-17, -1.3814992387869595e-17, 3.918434204559376e-17, -5.7771786349272854e-18, -9.042540472060099e-18, 9.293722151839546e-17, 1.1303175590075123e-17] 2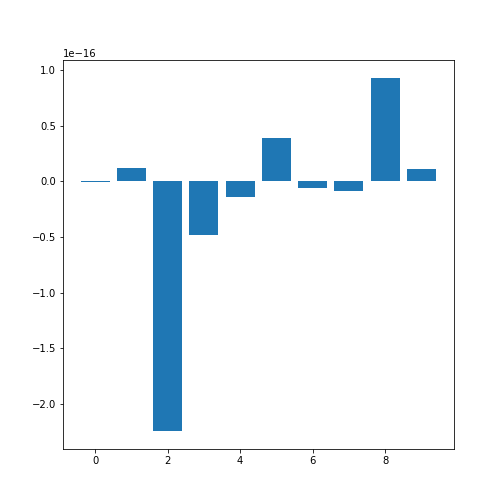
Note how the bars in this plot go up and down. The vertical axis, however, has values ranging from -10(-16) to +10(-16). This means that, for all practical purposes, all means are zero which is not a coincidence. The original values have been normalised to mean zero for the purpose of applying a machine learning algorithm to them.
In this example, we can clearly observe the importance of checking the data before working with them.
Exercises
End of chapter Exercises
Download the cervical cancer dataset provided, import it using
read_csv.
How many rows and columns are there?
How many columns contain floating point numbers (type float64)?
How many of the subjects are smokers?
Calculate the percentage of smokers
Plot the age distribution (with, for instance, 50 bins)
Get the mean and standard distribution of age of first sexual intercourse
Key Points
- Pandas package contains useful functions to work with DataFrames.
- The iloc property is used to index and slice a DataFrame.
- describe function is used to obtain a statistical summary of basic data features.
- The simplest method for data visualisation, is to use Pandas’ in-built functionality.
-
Matplotlibis a comprehensive library for creating static, animated, and interactive visualizations, in Python.